Lexagene Holdings (LXG.V) successfully used its MiQLab system to detect a group of common microbial contaminants called mycoplasmas.
These contaminants are responsible for substantial losses in time and money for biopharmaceutical companies, and finding them is the first step in dealing with them.
“We are increasingly harnessing the power of our MiQLab to drastically shorten the time to result for microbial biopharmaceutical contamination detection. Last month, Lexagene announced it could detect cutibacterium acnes 36 to 168 times faster than conventional methods. Through our research and development efforts, MiQLab technology can now readily detect mycoplasmas, a group of pervasive and highly problematic contaminants. We expect the time benefit of MiQLab mycoplasma test to be up to 300 times faster than conventional methods. These findings are significant for biopharmaceutical manufacturers and other life science companies. We are aggressively developing more tests for this industry so we can provide them a complete solution for their contamination testing needs,” said Dr Jack Regan, CEO and founder of Lexagene.
Lexagene is a healthcare tech company that develops molecular diagnostic systems for pathogen detection and genetic testing for molecular markers for on-site rapid testing for veterinary diagnostics and food safety. The markets these serve as clinical research, agricultural testing, and biodefence.
What are bioprocessing and mycoplasmas
Bioprocessing is using living organisms to make biological-based products. Brewing beer is a form of bioprocessing, as are vaccines and such transgenic scientific miracles like the oncomouse. The workflow can be broken down into three stages: upstream, harvest and downstream.
Upstream involves setting up culture conditions to maximize cellular growth. Once this has been achieved, they are harvested so the biological product can be extracted and worked on to make the final product.
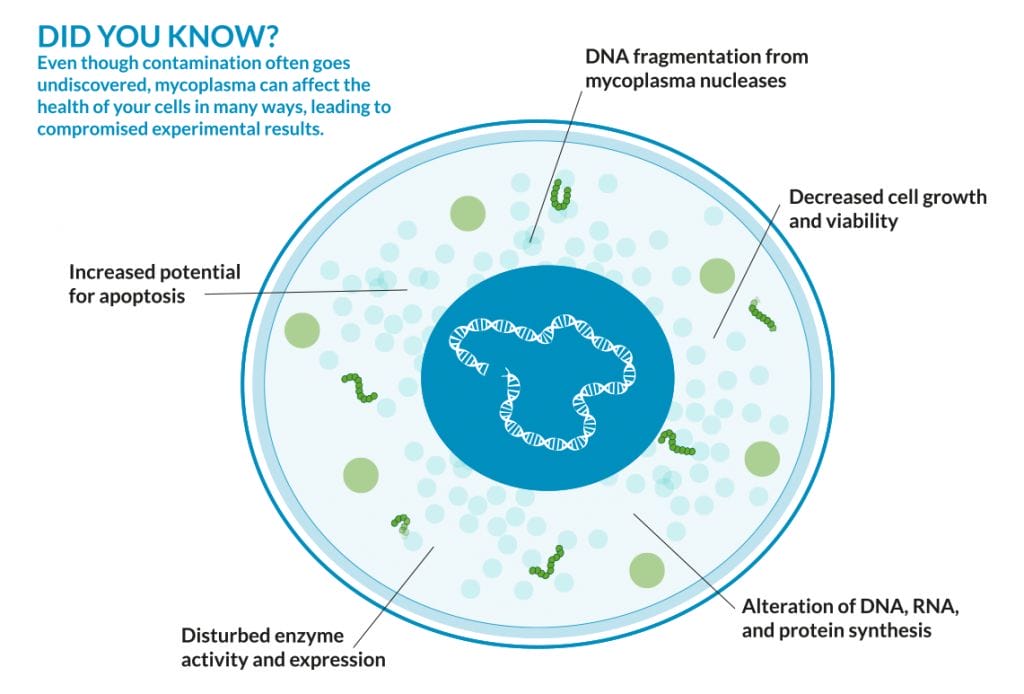
Mycoplasmas are a type of bacteria that lack a cell wall around their cell membranes. This makes them resistant to certain types of antibiotics (namely, those that target cell walls). They can be either parasitic (meaning, they feed off their host) or saprotrophic, which means they digest and eat whatever their target. Several species are particularly bad for humans, including M. Pneumoniae, which causes a condition commonly called ‘walking’ pneumonia and other respiratory disorders.
They’re also thought to contaminate 15 to 80% of cell cultures worldwide. Common methods for detection are time consuming, taking 28 days for definitive results. Routine testing during bioprocessing is required to maintain control over product yield, quailty, safety adn efficacy and to adequately determine if a cell is contaminated, and reducing the time is imperative.
Lexagene’s MiQLab mycoplasma test finds the most relevant mycoplasma species for microbial testing in bioprocessing, and more specifically the species Mycoplasma, arginini, mycoplasma orale, mycoplasma hyohinis, mycoplasma fermentans and acholeplasma laidlawii.
Lexagene’s results show that the MiQLab system can serve in a pinch as an alternative to lengthy traditional methods to detect mycoplasmas. It takes on average about two hours rather than the traditional test, which takes almost a month, and gets a definitive result. That’s a significant time advantage for biopharma manufacturers.
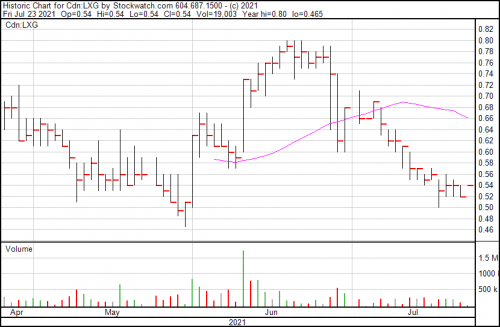
Lexagene is up a penny today and presently trading at $0.54.
—Joseph Morton



Great article! Follow me on Twitter @Guy_Leblanc . I’m LexaGene’s biggest fan and all in! Game changing technology that will change diagnostics at the point of care.